Research
Epigenetic regulation in meiotic recombination
Meiotic recombination, which refers to the exchange of genetic information between homologous chromosomes during meiosis, is an important source of genetic variation and thereby drives the evolution of sexual-reproductive organisms. Aberrant recombination is a major contributor to aneuploidy in mammals including human. Recombination is a complex process that is regulated by various factors, such as DNA sequence specificity, DNA methylation, nucleosome positioning, histone modification, chromatin accessibility, and 3D genome. The molecular mechanism of recombination still remains elusive in many aspects and we focus on: (1) the dynamics of chromatin structure, epigenetic signals and 3D genome in the process of recombination; (2) recombination-associated gene regulation network; (3) identification of hotspots using machine learning models; (4) the evolution of genome under the force of recombination.
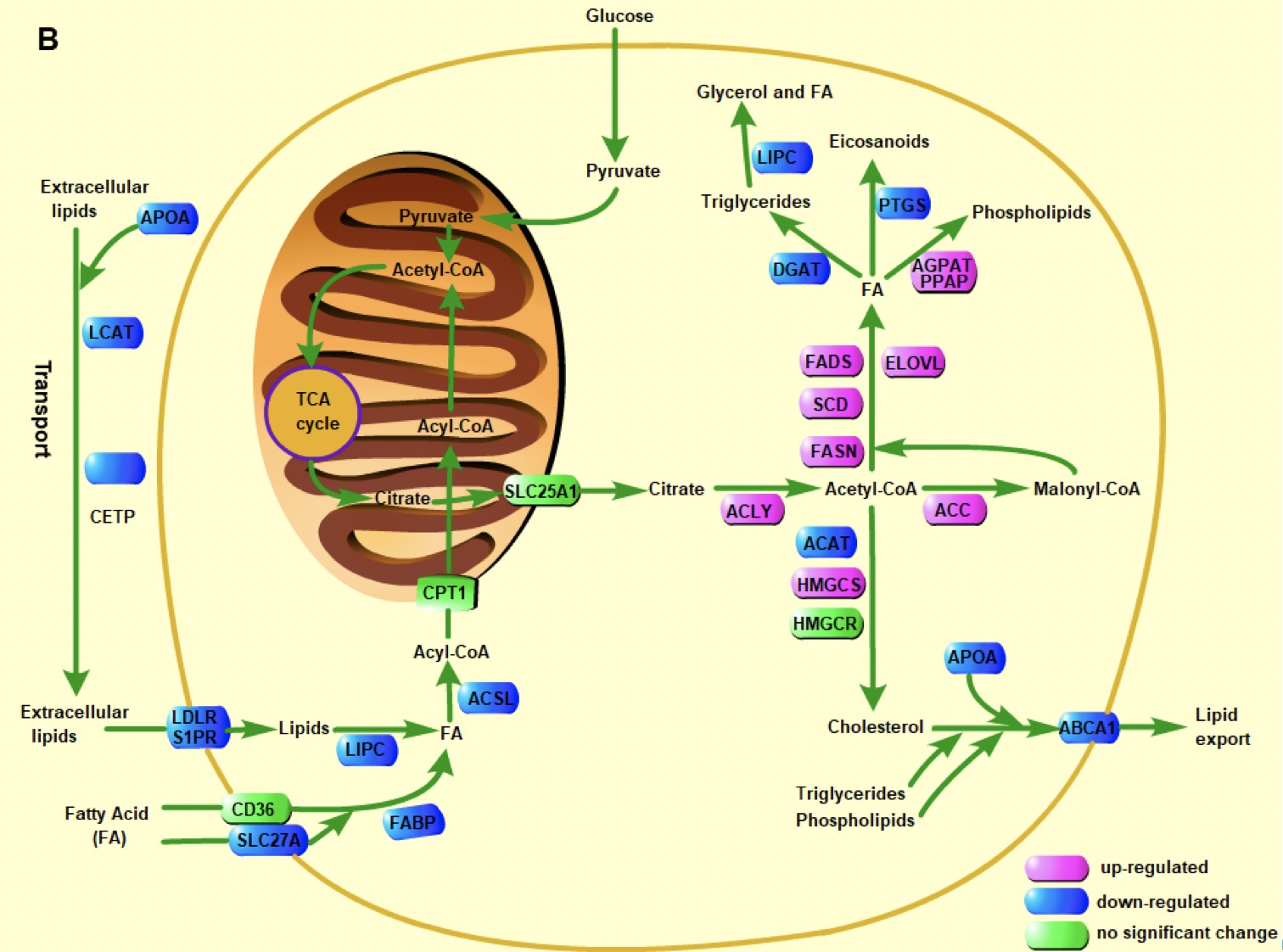
Metabolic reprogramming in cancer
In addition to the molecular level factors like genome variation, epigenomic signals and signaling pathways, metabolic reprogramming is believed to play an essential role in cancer development, but the exact molecular mechanism remains unclear. The energy metabolism, lipid metabolism and amino acid metabolism show dramatic difference between cancer and normal tissues. We are interested in investigating inter/intra-tumor metabolic heterogeneity and the dynamics of metabolic pathways during cancer development.
Modeling chromatin structure
Chromatin structure can be understood at multi-level: nucleosome particle, which is the basic structural unit of chromatin and composed of DNA double helix wrapped around histone octamer, 30 nm chromatin structure, which is a product of further condensation of nucleosome arrays, loop structure, and the most densely packaged structure formed in metaphase. The packaging rule of chromatin regulates diverse fundamentally important processes, such as gene transcription, DNA replication and meiotic recombination, and consequently affects cell differentiation and development. We are interested in deciphering the rule of nucleosome sliding, modeling 30 nm structure, and 3D modeling of chromatin based on Hi-C map.