Publications
Highlights
(For a full list see below or go to Google Scholar)
Guoqing Liu*, Guo-Jun Liu, Xiangjun Cui, Ying Xu
Frontiers in Cell and Developmental Biology 2020
Full List
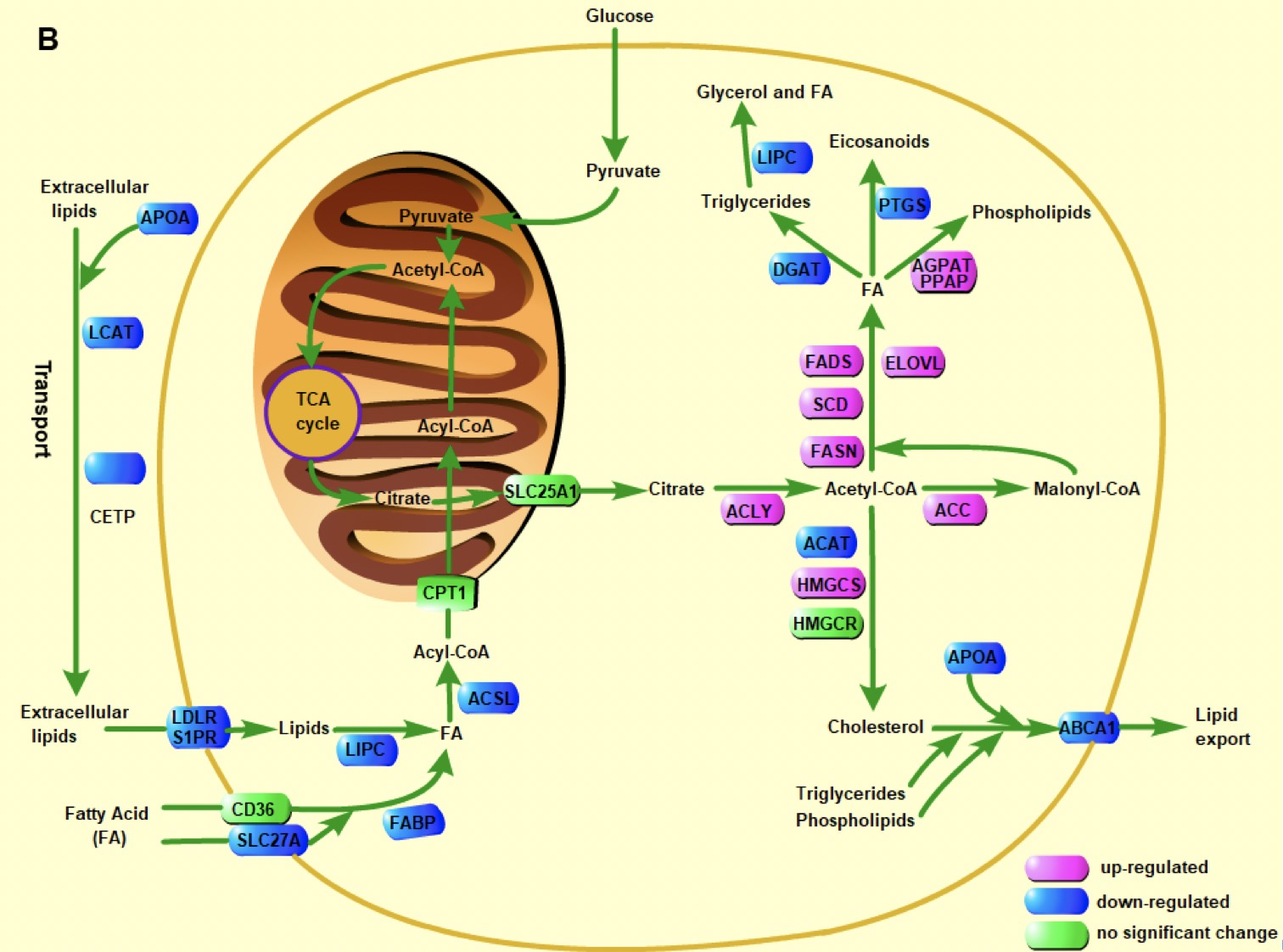
Transcriptomic data analyses reveal a reprogrammed lipid metabolism in HCV-derived hepatocellular cancer
Guoqing Liu*, Guo-Jun Liu, Xiangjun Cui, Ying Xu
Frontiers in Cell and Developmental Biology 2020
Dynamic Alternative Splicing During Mouse Preimplantation Embryo Development
Xing Y, Yang W, Liu G, Cui X, Meng H, Zhao H, Zhao X, Li J, Liu Z, Zhang MQ, Cai L.
Frontiers in Bioengineering and Biotechnology 2020
A co-expression network for differentially expressed genes in bladder cancer and a risk score model for predicting survival
Chen Z, Liu G, Hossain A, Danilova IG, Bolkov MA, Liu G, Tuzankina IA, Tan W.
Frontiers in Bioengineering and Biotechnology 2020
DNA physical properties outperform sequence compositional information in classifying nucleosome-enriched and -depleted regions
Guoqing Liu*, Guo-Jun Liu, Jiu-Xin Tan, Hao Lin
Genomics 2019
Identification of miR-200c and miR141-mediated lncRNA-mRNA crosstalks in muscle-invasive bladder cancer subtypes
Guojun Liu, Zihao Chen, Irina G. Danilova, Mikhail A. Bolkov, Irina A. Tuzankina, Guoqing Liu*
Frontiers in Genetics 2018
The affinity of DNA sequences containing R5Y5 motif and TA repeats with 10.5-bp periodicity to histone octamer in vitro
Hongyu Zhao, Fenghui Zhang, Mingxing Guo, Yongqiang Xing, Guoqing Liu, Xiujuan Zhao, Lu Cai*
J Biomol Struct Dyn, 2018
The implication of DNA bending energy for nucleosome positioning and sliding
Guoqing Liu*, Yongqiang Xing, Hongyu Zhao, Lu Cai, Jianying Wang
Scientific Reports 2018
Physical properties of DNA may direct the binding of nucleoid-associated proteins along the E. coli genome
Guoqing Liu, Qin Ma, Ying Xu
Mathematical Biosciences 2018
iRSpot-Pse6NC: Identifying recombination spots in Saccharomyces cerevisiae by incorporating hexamer composition into general PseKNC
Hui Yang, Wang-Ren Qiu, Guoqing Liu, Feng-Biao Guo, Wei Chen, Kuo-Chen Chou, Hao Lin*
International Journal of Biological Sciences, 2018
Evolutionary direction of processed pseudogenes
Guoqing Liu*, Xiangjun Cui, Hong Li, Lu Cai
Science China Life Sciences, 2016
A deformation energy-based model for predicting nucleosome dyads and occupancy
Guoqing Liu, Yongqiang Xing, Hongyu Zhao, Jianying Wang, Yu Shang and Lu Cai
Scientific Reports, 2016
MiasDB: A Database of Molecular Interactions Associated with Alternative Splicing of Human Pre-mRNAs
12. Yongqiang Xing, Xiujuan Zhao, Tao Yu, Dong Liang, Jun Li, Guanyun Wei, Guoqing Liu, Xiangjun Cui, Hongyu Zhao, Lu Cai*
Plos One 2016
Nucleosome organization around pseudogenes in the human genome
Guoqing Liu, Fen Feng, Xiujuan Zhao, Lu Cai
BioMed Research International, 2015
GAA triplet-repeats cause nucleosome depletion in the human genome
Hongyu Zhao, Yongqiang Xing, Guoqing Liu , Ping Chen, Xiujuan Zhao, Guohong Li, Lu Cai*
Genomics. 2015
Using weighted features to predict recombination hotspots in Saccharomyce scerevisiae
Guoqing Liu, Yongqiang Xing, Lu Cai
Journal of Theoretical Biology, 2015
Predicting recombination hotspots in yeast based on DNA sequence and chromatin structure
Bingjie Zhang, Guoqing Liu*
Curr Bioinformatics, 2014
Compositional bias is a major determinant of the distribution pattern and abundance of palindromes in Drosophila melanogaster
Guoqing Liu, Jia Liu, Bingjie Zhang
J Mol Evol, 2012
Calculation of nucleosomal DNA deformation energy: its implication for nucleosome positioning
Jian-Ying Wang, Jingyan Wang, Guoqing Liu*
Chromosome Research, 2012
Sequence-dependent prediction of recombination hotspots in Saccharomyces cerevisiae
Guoqing Liu*, Jia Liu, Xiangjun Cui, Lu Cai
J Theor Biol, 2012
Processed pseudogenes are located preferentially in regions of low recombination rates in the human genome
Guoqing Liu*, Hong Li, Lu Cai
J Evol Biol, 2010
The correlation between recombination rate and dinucleotide bias in Drosophila melanogaster
Guoqing Liu, Hong Li*
Journal of Molecular Evolution, 2008
假基因研究进展
刘国庆,白音宝力高,邢永强
生物化学与生物物理进展,2010
人类基因组信息量的扩增速率受重组的影响
刘国庆,罗辽复
生物化学与生物物理进展,2012
Genome-wide characterization and prediction of Arabidopsis thaliana replication origins
Yong-qiang Xing, Guo-qing Liu, Xiu-juan Zhao
BioSystems 124 (2014)
Correlations between recombination rate and intron distributions along chromosomes of C. elegans
Hong Li, Guoqing Liu, Xuhua Xia
Progress in Natural Science, 2009
The effect of impurities in a three-qubit Ising spin Model
Jia Liu, Guo-Qing Liu, Xiao-Hong Du
International Journal of Theoretical Physics, 2012
An analysis and prediction of nucleosome positioning based on information content
Xing Yong-qiang, Liu Guo-qing, Zhao Xiu-juan, Cai Lu*
Chromosome Res, 2013
Combinatorial patterns of histone modifications in Saccharomyces.cerevisiae
Xiang-Jun Cui, Hong Li, Guo-Qing Liu
Yeast, 2011
Predicting nucleosome binding motif set and analyzing their distributions flanking functional sites of human genes
Tonglaga Bao, Hong Li, Xiaoqing Zhao, Guoqing Liu
Chromosome Research, 2012
Pre-mRNA选择性剪接的调控及选择性剪接数据库
邢永强,刘国庆,蔡禄
中国生物化学与分子生物学报, 2016
拟南芥种子萌发过程中差异表达基因的识别和pre-mRNA可变剪接图谱的构建
邢永强,王延新, 何泽学,崔向军,刘国庆, 孟虎, 赵宏宇,赵秀娟,蔡禄
科学通报,2019
拟南芥不同组织基因表达及可变剪接差异分析
邢永强,赵宏宇,刘国庆,赵秀娟,蔡禄
生物物理学报,2014